Drug–target interaction (DTI) prediction is a crucial step in drug discovery and repositioning as it reduces experimental validation costs if done right. Thus, developing in-silico methods to predict potential DTI has become a competitive research niche.
LM-DTI is a web server for drug–target interactions prediction. It is based on the heterogeneous network data fusion,network embedding and ensemble learning. It can serve the following functions.
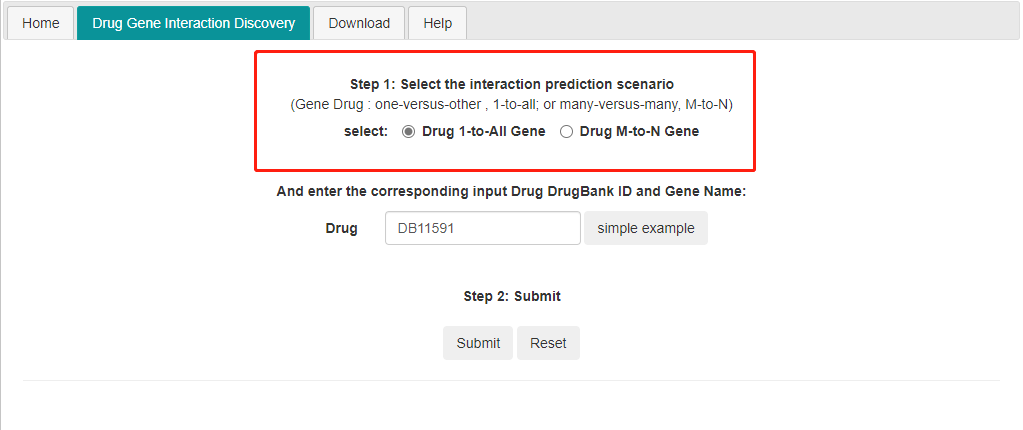
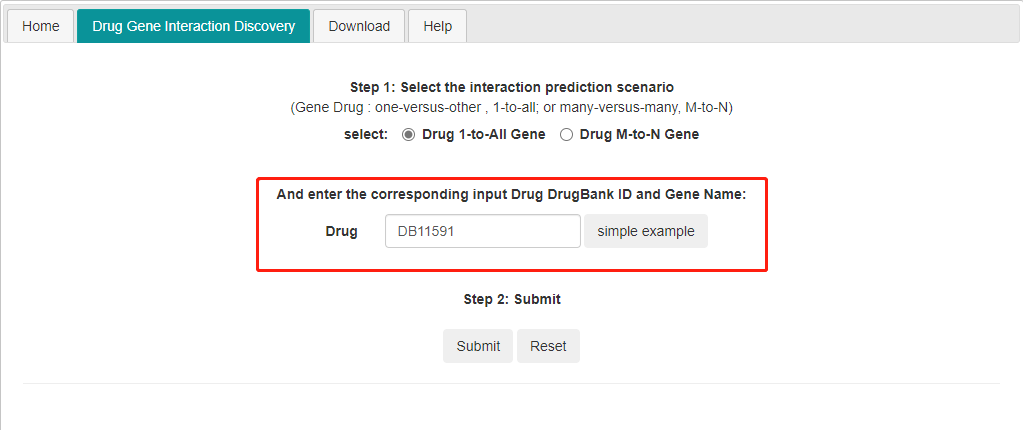
1) 1 to all DTI prediction. Input one drug id and it predicts the interaction between the input drug and all the genes in the database.
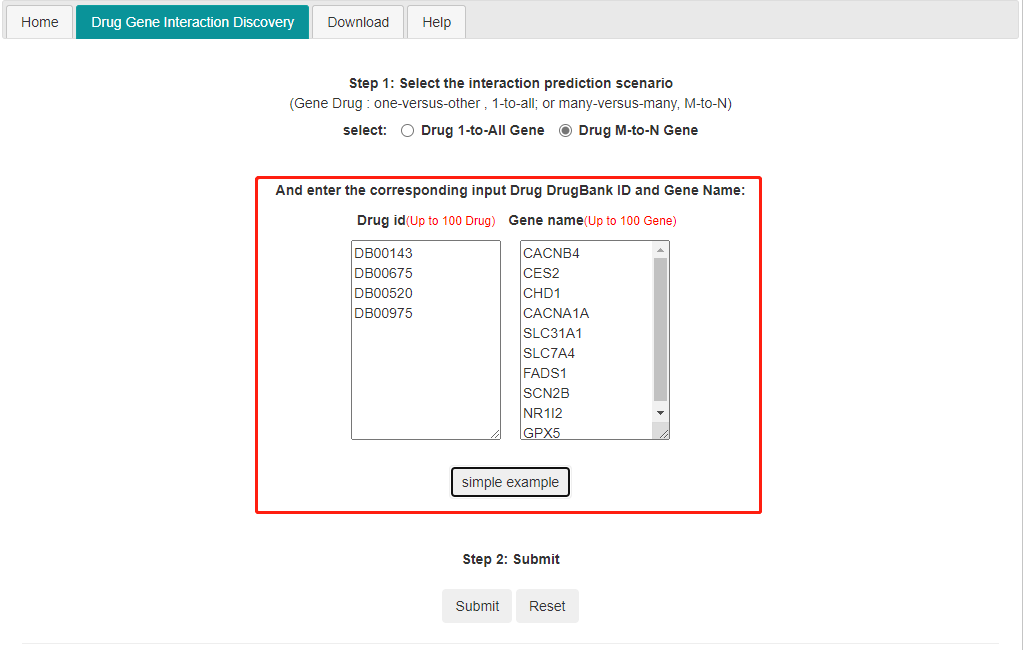
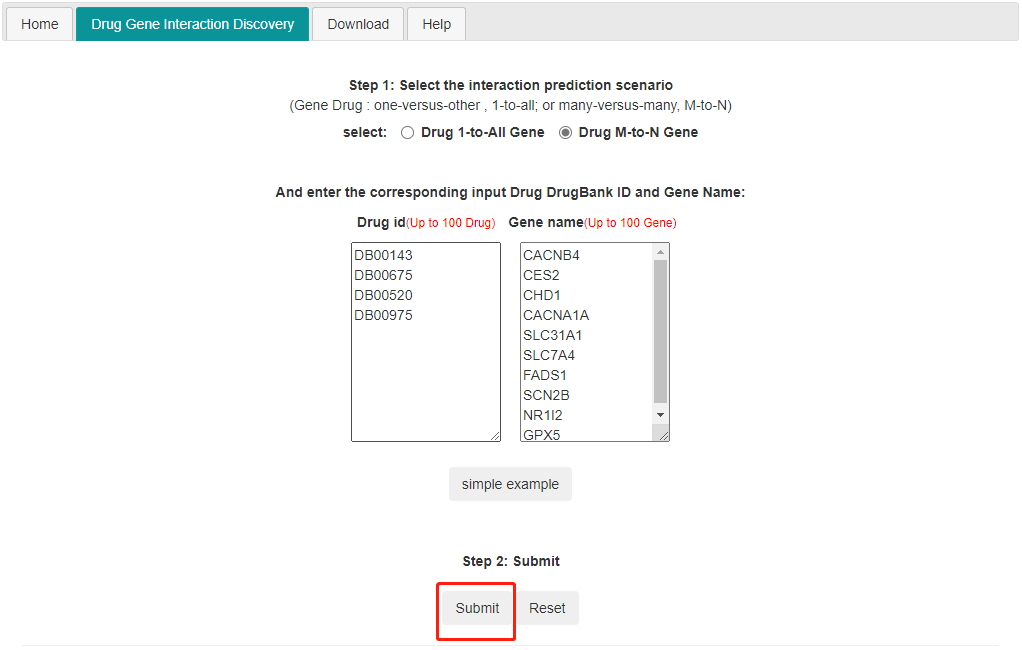
2) M to N DTIprediction. Input m drug and n genes(up to 100) and it predicts the interaction between the input drug and the input genes.
All analyses are performed at the levels of heterogeneous network topology. The full prediction datasets can also be downloaded in the "Download" page.
Comprehensive usage instructions can be found in the "Help" menu.
Jul 2018: the original LnCompare program is released.
Feb 2019: a stable similar gene discovery module has been established.
Dr. Jianwei Li, school of artificial intelligence, Hebei University of Technology, Tianjin 300401, China
Email: lijianwei@hebut.edu.cn
LM-DTI works on DrugBank id , not drug name.
This website has been tested by using Chrome, Microsoft Edge and Firefox browsers. Microsoft IE may not work well.
Citation: Jianwei Li, Zhiguang Li, Yinfei Wang, Hongxin Lin, Baoqin Wu. LM-DTI:a tool of predicting drug-target associations by node2vec and network path score method.
Download the entire prediction reault for LM-DTi here: table_prediction.
Note: If users choose the M-to-N prediction, the maximum size of each set is 100.

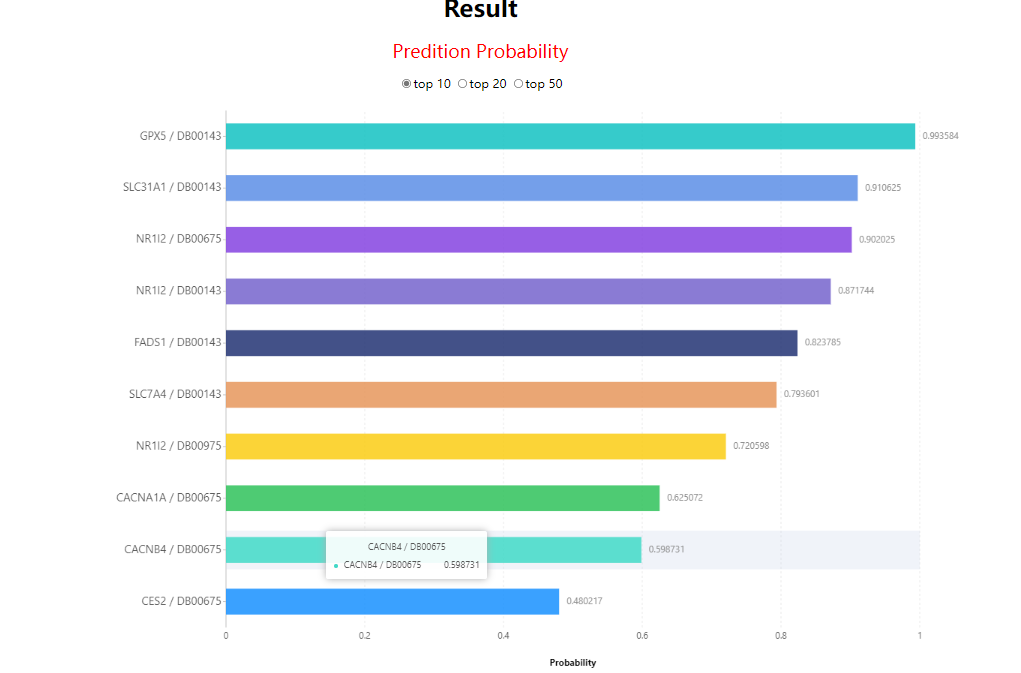
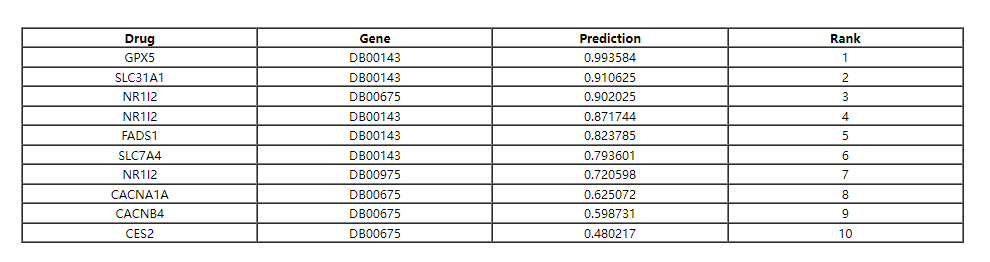
The entire reults table of LM-DTI can be found on the download page, user can click 'table_prediction' link to download it.
